In Riyadh I spent a rainy afternoon trying to work out how this was done (no, not really rainy but stayed indoors instead of driving about the desert). There's not much on the web about it. For at least a year people have been saying there will be an app - and yes, one will eventually appear, but for now this is how we can get our kicks.
rwiki says that it's easy to install R on Android but it needs a lot of patching (so perhaps not so easy after all).
At LinkedIn (may need to subscribe to see this) a couple of clues from clever geeks said things like:
"...Just set up an Rstudio server using an Amazon instance and be done as you can use it from any internet connection"
and:
"You could run r in an amazon aws instance (a micro instance is free right now for limited hours / month) and access that from a tablet or computer if you use r studio server. The advantages are that - works on any platform..."
So how can you do this on Amazon AWS (Amazon Web Services)? It must be so easy but the geeks don't say - and it's free. Never having used these services before, one becomes easily overwhelmed by the sheer number of services on offer - which one to choose? See: http://aws.amazon.com/free/
I set it all up on my laptop first (running Windows).
You should choose Amazon EC2 (Amazon Elastic Compute Cloud). You will need to click on the Get Started for Free button, you need to register a credit card.
Once you are registered you should be able to select a console page. If not, go here:https://console.aws.amazon.com/console/homeand click on the icon for EC2
This gives me the EC2 dashboard. For some reason I have been allocated resources at US West (Oregon), and yet I am in Saudi Arabia. Hopefully you will find a nearer more appropriate allocation (or maybe it just doesn't matter). From here you want to Launch Instance.
You will find the instructions in this youtube video most helpful for the rest of setting up the EC2:http://www.youtube.com/watch?NR=1&v=OLfmqcYnhUM
A floating window appears Create a New Instance. Click Continue (Classic Wizard).
A dazzling range of AMIs (Amazon Machine Images) are available. I selected the Microsoft Windows Server 2008 Base 64 bit (starred indicating it is free).
As suggested in the video select the default settings.
From the Windows Start button search for Remote Desktop Connection to launch the server window. From the floating window that comes up when you connect, you should copy & paste the DNS address (this is different each time you make a connection). You need to type in Administrator and the generated password - as seen in the video (generating the password may take a while, 15 or more minutes, keep refreshing to check).
Once you are in to the Windows server run Explorer and install R from CRAN.
While in Windows I found it useful to change my password to something more memorable.When finished, Logout. Back in normal Windows you should Stop the instance. Your usage will then stop. Important: do not Terminate or this will remove the instance completely (R installation and all).
Go to android phone.
Install a remote connection client app, I tried 2X Client RDP/Remote Desktop which works very well and is free.
Update:
It is well worth installing the AWS Console app on the phone. This makes it much less fiddly stopping and starting the instance. For some reason the default is for Virginia, so you may need to change the area.
Run web browser. Go to the EC2 Management Console page, as seen earlier on laptop. You should see the instance that you set up, and it should be in a stopped state (assuming you stopped it earlier).Select the instance, and from the Actions menu click on Start, confirm Yes on floating window.It will show pending and changing quite soon to show running.From the Actions menu select Connect. This will give you the login information and you need to copy and paste the Public DNS from here.
Run the 2X Client, choose an RDP connection, then:
1. paste Public DNS into the Server
2. enter Administrator for name (if local network specifies its own domain then enter amazon\Administrator instead)
3. enter password from before
4. Select Connect to Console
5. Connect, click on icon next to X, then select item
After a few seconds delay, you should see the screen below (as on my Samsung Galaxy SIII). There is a dinky mouse with pointer (shown in center of screen) and a keyboard. Extra characters are available by selecting Shift.
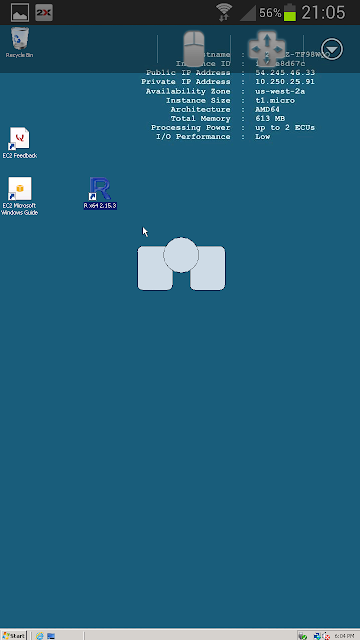
Running R gives the screen below. Very small and fiddly - but definitely worth it!
Once you have logged out, remember to stop the instance else you could run up charges (it may be hard to find the floating window asking for Yes confirmation of this).
After this my usage report (downloaded CSV) looked massive...But Account activity = £0.0
rwiki says that it's easy to install R on Android but it needs a lot of patching (so perhaps not so easy after all).
At LinkedIn (may need to subscribe to see this) a couple of clues from clever geeks said things like:
"...Just set up an Rstudio server using an Amazon instance and be done as you can use it from any internet connection"
and:
"You could run r in an amazon aws instance (a micro instance is free right now for limited hours / month) and access that from a tablet or computer if you use r studio server. The advantages are that - works on any platform..."
So how can you do this on Amazon AWS (Amazon Web Services)? It must be so easy but the geeks don't say - and it's free. Never having used these services before, one becomes easily overwhelmed by the sheer number of services on offer - which one to choose? See: http://aws.amazon.com/free/
I set it all up on my laptop first (running Windows).
You should choose Amazon EC2 (Amazon Elastic Compute Cloud). You will need to click on the Get Started for Free button, you need to register a credit card.
Once you are registered you should be able to select a console page. If not, go here:https://console.aws.amazon.com/console/homeand click on the icon for EC2
This gives me the EC2 dashboard. For some reason I have been allocated resources at US West (Oregon), and yet I am in Saudi Arabia. Hopefully you will find a nearer more appropriate allocation (or maybe it just doesn't matter). From here you want to Launch Instance.
You will find the instructions in this youtube video most helpful for the rest of setting up the EC2:http://www.youtube.com/watch?NR=1&v=OLfmqcYnhUM
A floating window appears Create a New Instance. Click Continue (Classic Wizard).
A dazzling range of AMIs (Amazon Machine Images) are available. I selected the Microsoft Windows Server 2008 Base 64 bit (starred indicating it is free).
As suggested in the video select the default settings.
From the Windows Start button search for Remote Desktop Connection to launch the server window. From the floating window that comes up when you connect, you should copy & paste the DNS address (this is different each time you make a connection). You need to type in Administrator and the generated password - as seen in the video (generating the password may take a while, 15 or more minutes, keep refreshing to check).
Once you are in to the Windows server run Explorer and install R from CRAN.
While in Windows I found it useful to change my password to something more memorable.When finished, Logout. Back in normal Windows you should Stop the instance. Your usage will then stop. Important: do not Terminate or this will remove the instance completely (R installation and all).
Go to android phone.
Install a remote connection client app, I tried 2X Client RDP/Remote Desktop which works very well and is free.
Update:
It is well worth installing the AWS Console app on the phone. This makes it much less fiddly stopping and starting the instance. For some reason the default is for Virginia, so you may need to change the area.
Run the 2X Client, choose an RDP connection, then:
1. paste Public DNS into the Server
2. enter Administrator for name (if local network specifies its own domain then enter amazon\Administrator instead)
3. enter password from before
4. Select Connect to Console
5. Connect, click on icon next to X, then select item
After a few seconds delay, you should see the screen below (as on my Samsung Galaxy SIII). There is a dinky mouse with pointer (shown in center of screen) and a keyboard. Extra characters are available by selecting Shift.

Running R gives the screen below. Very small and fiddly - but definitely worth it!
Once you have logged out, remember to stop the instance else you could run up charges (it may be hard to find the floating window asking for Yes confirmation of this).
After this my usage report (downloaded CSV) looked massive...But Account activity = £0.0
Perhaps on another rainy afternoon I'll set up RStudio for my phone.







